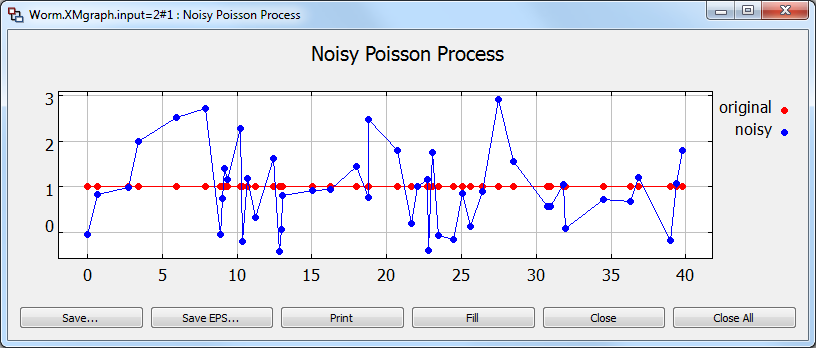
Now you can execute the simulation but bear in mind that both PTcl and parameter file have to be at the same location. The figures below show the model representation within the MLDesigner GUI and the result of the simulation.
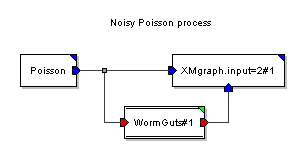
An example of a simulation that contains both an SDF part and a DE part.
In this example, a Poisson process where particles have value 0.0 is sent into an SDF wormhole, where Gaussian noise is added to the samples. This demo shows how easy it is to use the SDF primitives to perform computation on DE particles. The overall delay of the SDF wormhole is zero, so the result is simply Poisson arrivals of Gaussian noise samples.
A wormhole has an outer domain and an inner domain. The outer domain is determined by the current domain at the time you start the defmodule command to create the wormhole. The inner domain is determined by the domain command that appears inside the module definition.
reset
domain DE
# importing libraries for data structure definition
source $MLD/MLD_Libraries/SDF/NumberGenerators/NormalFloatConst/NormalFloatConst.ptcl
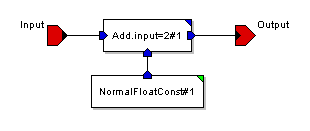
defmodule wormGuts {
domain SDF
# definition of model parameters
newparam stopTime float {$stopTime}
newparam curIter int {$curIter}
newparam absIter int {$absIter}
newparam absSimul int {$absSimul}
# definition of instances and their properties
instance Add.input=2#1 AddFloat
numports Add.input=2#1 Input 2
instance NormalFloatConst#1 NormalFloatConst
trysetparam NormalFloatConst#1 {Mean} {0}
trysetparam NormalFloatConst#1 {Variance} {1}
trysetparam NormalFloatConst#1 {Seed} {-1}
# define the connections
alias Output Add.input=2#1 Output
alias Input Add.input=2#1 Input#1
connect NormalFloatConst#1 Normal Add.input=2#1 Input#2
}
defsystem worm DE
# define the target and set the target parameters
target default-DE
trytargetparam timeScale {1.0}
trytargetparam usedScheduler {0}
# definition of model parameters
newparam {GlobalSeed} {int} {1234567890}
newparam {RunLength} {int} {40}
# definition of instances and their properties
instance WormGuts#1 wormGuts SDF
instance Poisson Poisson_2_3
trysetparam Poisson {meanTime} {1.0}
trysetparam Poisson {magnitude} {1.0}
instance XMgraph.input=2#1 XMgraph
numports XMgraph.input=2#1 input 2
trysetparam XMgraph.input=2#1 {title} {Noisy Poisson Process}
trysetparam XMgraph.input=2#1 {saveFile} {}
trysetparam XMgraph.input=2#1 {options} {-P -0 original -1 noisy =800x300+0+0}
trysetparam XMgraph.input=2#1 {Cumulation} {0}
# define the connections
node node1
nodeconnect Poisson output node1
nodeconnect WormGuts#1 Input node1
nodeconnect XMgraph.input=2#1 input#1 node1
connect WormGuts#1 Output XMgraph.input=2#1 input#2
# execution of iterations
if {[info exists parameterfile] &&
[info exists parameterset] &&
[info exists parameteriter]} {
simulate $parameterfile $parameterset $parameteriter
} else {
puts "Cannot execute simulation without specification of the parameter file."
puts "One possible reason is that you tried to simulate the system using \"source\""
puts "command. Use \"execute\" command instead to run simulations, see \"help execute\""
}
delsystem worm
wormhole_example.ptcl
Save this code in a file named wormhole_example.ptcl. Before you can execute this example, you need to create a parameter file (*.params). Its name has to be consistent with the name of the PTcl file, in this case wormhole_example.params.
PARAMETER_SET
{
GlobalSeed : 1234567890
RunLength : 40
}
wormhole_example.params
Now you can execute the simulation but bear in mind that both PTcl and parameter file have to be at the same location. The figures below show the model representation within the MLDesigner GUI and the result of the simulation.